Scientific Facilities
Fish
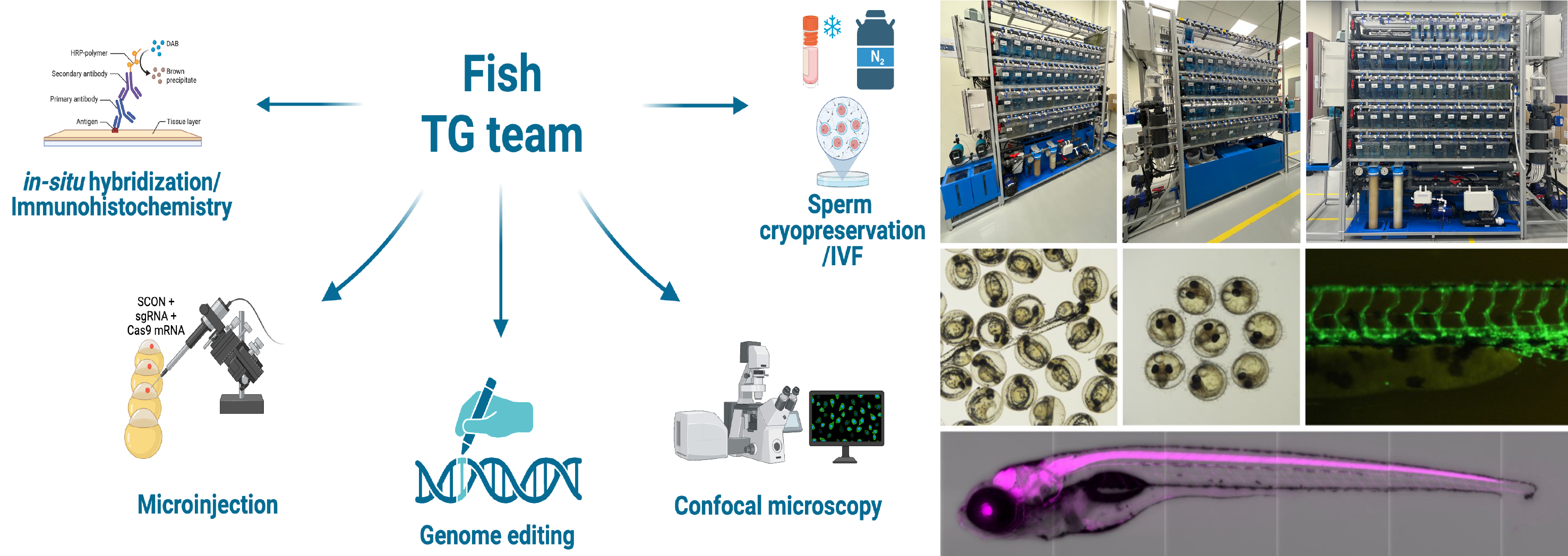
Borderless gene editing in fish
Applying precise and efficient gene editing technologies, such as CRISPR/Cas9 or HDR-based methods, can significantly improve the accuracy and success rate of genetic research. Studying a wide range of fish species with different developmental characteristics allows researchers to optimize editing strategies and expand the practical applications of gene editing across diverse biological contexts. Considering the importance of this work, the CGE established a Fish Facility, where we are working on the following projects:
- Project 1. Optimization and generation of conditional knockout fish utilizing the SCON (Short Conditional IntrON) technique
Due to embryonic lethality, the study of developmentally essential genes is limited to the early stages. This issue can be resolved through the generation of a conditional knock-out for essential genes. The SCON technique (Wu et al., 2022) is a universal conditional intron system for a conditional knock-out approach applicable to various animal models, such as zebrafish and medaka. By generating SCON-integrated fish, we can investigate the functions of essential genes spatially and temporally.
- Project 2. Generation of transgenic and mutant fish using the CRISPR/Cas9 system
Using a CRISPR/Cas9 system and applying microinjection and electroporation methods, we will first identify genes of interest, including those involved in sperm-egg recognition, cold-resistance, and disease-resistance, in zebrafish and medaka, and then apply our findings to other fish species. For example, applying gene editing in marine fish can significantly shorten the time required to develop higher-quality stocks compared to conventional selective breeding methods.
- Project 3. Establishment of
intestinal organoids from fish
Studying bacterial- and virus-infection diseases in farmed fish has been challenging due to the long generation time and limited tools being available. To overcome these limitations, our goal is to establish intestinal organoids from fish and investigate diseases in farmed fish.
Contact : Jung-Hwa Choi (jhchoi@ibs.re.kr)

